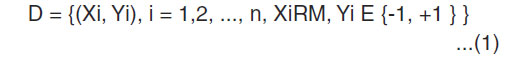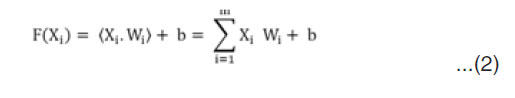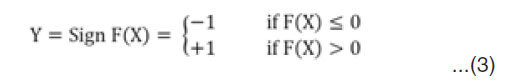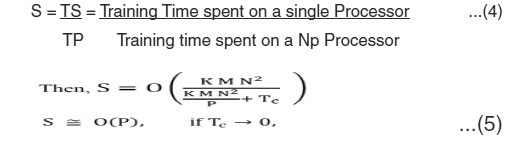Rabie A. Ahmed1 and Mohammed M. Al-Shomrani2
1Northern Border University, King Abdulaziz University, Faculty of Science,Saudi Arabia.
2Faculty of Computing and IT, Rafha, KSA, P. O. Box 80203, Jeddah 21589, Saudi Arabia.
Article Publishing History
Article Received on :
Article Accepted on :
Article Published : 18 Apr 2015
Article Metrics
ABSTRACT:
Support Vector Machine SVM is one of the most effective Machine Learning Algorithms for data classification, which has a significant area of research. Since the training process of large data sets is very computationally intensive, there is a need to improve its efficiency using high performance computing techniques. Initially, SVM was used for binary classification, but most applications have more than two categories, resulting in multiclass classification. Classifying different categories among large data sets has become one of the most important computing problems. Since the complexity of training of non-linear SVMs has been estimated to be quadratic in the number of training examples, which is unreasonable for data sets with tens of thousands of training examples. In this paper, we developed an efficient parallel algorithm which combines Parallel Binary Class with Parallel Multi Class Support Vector Machines for classification, PMC-PBC-SVM, which means two level parallel algorithm. The main idea of this algorithm is how to divide a set of processors into two subsets, one is responsible for the multi class case and the other is responsible for the binary class case. PMC-PBC-SVM algorithm was implemented using C++ programming language and Message passing Interface, MPI, communication routines. The parallel code was executed on an ALBACORE Linux cluster, and then tested with four data sets with difference sizes, Earthworm, Protein, Mnist, and Mnist8m. The results show that the PMC-PBC-SVM implementation can significantly improve the performance of data classification without loss of accuracy. The results also demonstrated a form of proportionality between the size of the data set and the PMC-PBC-SVM efficiency. The larger the data set is the higher efficiency the PMC-PBC-SVM achieves.
KEYWORDS:
Classification; SVM; Data sets; Parallel computing; MPI
Copy the following to cite this article:
Ahmed R. A, Al-Shomrani M. M. Two Level Parallelism Implementation to Classify Multi class Large Data Sets. Orient.J. Comp. Sci. and Technol;7(3)
|
Introduction
Databases have a lot of hidden information that is useful for making intelligent decisions. There are many methods that can be used to extract information from databases. Classification is an important technique for data analysis. Most algorithms assume a small data size, but recent database mining research has a large data set, therefore there is a need to develop these algorithms that handle large data sets. These methods often use parallel and distributed processing techniques to solve this problem1.
Support Vector Machine (SVM) is one of the most effective machine learning algorithms for
data classification, which has a significant area of research. The classification problem consists of two phases, training phase and testing phase. In the training phase, the training data set is used to train SVM model while the testing data set is used to test this model in the testing phase. Since the training process of large data sets is very computationally intensive, there is a need to improve the performance using high performance computing techniques. Initially, SVM was used for binary classification, i.e., for identifying two classes. Most applications have more than two categories, resulting in multi category classification. Multi class classification consists of identifying a class among k categories which increases the complexity by k times2.
The main idea of the SVM classification algorithm is to separate two-point classes of a training data set with a surface that maximizes the margin between them3. Therefore, for a training data set D, where
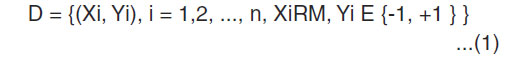
Which has data samples Xi and labels Yi, the objective is to find a function
F: RM – R, for trained parameters Wi and b,
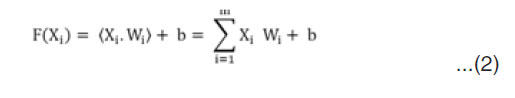
Then, for any sample (X,Y), the class assigned by the model is
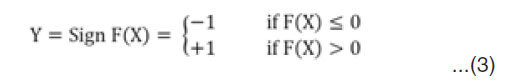
For Multi class SVM classification, the algorithm that performs the training is the One Versus One (OVO) algorithm which generates ( ) tasks to be executed concurrently among the available p processors where K is the number of classes in the training set. The decomposition technique used here is called, data decomposition, as different processing elements (PEs) perform similar tasks on different data partitions. The complexity of the multi class classification is ( ), where K is the number of classes, M is the number of features, and N is the number of training samples. This serial complexity is the worst case scenario for multi class classification using binary classifiers. But when this job is distributed among P processors, the parallel complexity becomes ( ) where is the complexity due to communication for task scheduling and combining the results. The parallel performance is evaluated by the relative speedup (S), which is defined as the ratio of the time taken to solve a problem on a single processing element to the time required to solve the same problem on a parallel computer with p identical processing elements. Also, Efficiency (E) is another way to analyze the parallel implementation, which is defined as the ratio of speedup to the number of processing elements4.
Related Work
One of the most common SVM solvers is LIBSVM5. The complexity of training of non-linear SVMs with solvers such as LIBSVM has been estimated to be quadratic in the number of training examples4, which can be unreasonable for data sets with hundreds of thousands of examples. Researchers have therefore explored ways to achieve faster training times. For linear SVMs very efficient solvers are available which converge in a time which is linear in the number of examples6. The LIBSVM5 software is developed for a working set of size two, which is tending to minimize the computational cost per iteration. Actually, in this case the inner Quadratic Programing, QP, sub-problem can be systematically solved without requiring a numerical QP solver and the updating of the objective gradient only involves the two Hessian columns corresponding to the two updated variables. On the other hand, if only few variables are updated per iteration, slow convergence is normally implied. The SV Mlight algorithm7 uses a more general decomposition strategy to exploit working sets of size larger than two. By updating more variables per iteration, such an approach is more suitable for a faster convergence, but it introduces additional difficulties and costs. Following the SVMlight decomposition techniques, another effort to reach a good way between convergence rate and cost per iteration was introduced in8. Unlike SVMlight, it is considered medium or large sized working sets, ( ) ( ), that allow the method to converge in a small number of iterations where the most costly tasks, such as sub problem solving and gradient updating, can be simply and effectively distributed between available processors. Based on this idea, a new parallel gradient projection-based decomposition technique (PGPDT) is developed and implemented in software to train support vector machines for binary classification problems in parallel as in 9. Authors in 10, combine ideas in 4 and 9 to develop an efficient parallel algorithm, SMC-PBC-SVM, which combines Parallel Binary Class with Serial Multi Class Support Vector Machines for classification. Also, a lot of work has been done in 11 and 12 to develop kernel computation costs. Also, other works are made in13,14, 15 and16 to optimize a working set size selection. On the other hand, authors in 17 were developed SVM training by quickly removing most of non-support vectors.
Motivation
In recent years, the word parallel has become one of the most used words in computer science research. Every area in computer science seems to be developed by it from parallel operating systems to parallel architectures to parallel algorithms to parallel languages and compilers. Rapid progress made in bioinformatics and other fields need a faster and a smarter machine learning algorithms for data classification. Therefore, classification of bioinformatics and large data sets, which needs a significant computational power, is an ongoing research area. Parallel processing is one of the most effective methods for the fast solution of computationally intensive problems. Parallel processing is making a tremendous impact in many areas of computer applications. A growing number of applications in science, engineering, business, and medicine are requiring computing speeds that hardly can be achieved by the current 3
conventional computers. The objective of our work is to develop a parallel algorithm, PMC-PBC-SVM, which combines Parallel Binary Class with Parallel Multi Class Support Vector Machines for classification. Then, implement this algorithm using C++ programming language and standard Message passing Interface MPI communication routines. After that, the parallel codes will execute on an ALBACORE Linux cluster and will test with different size data sets. Then, the performance results will analyze using speed up and efficiency. Finally, performance analysis will visualize using Jump shot software.
Pmc-Pbc-Svm Algorithm
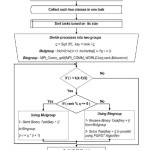
All previous results are encouraging, but more work needs to be done. In this section, a new classification algorithm is introduced. The new algorithm merges parallel binary classification with Parallel multi class classification to produce an efficient parallel algorithm for classification which is named PMC-PBC-SVM. The main idea in this algorithm is how to divide a set of processors into two subsets, one is responsible for the multi class case and the other is responsible for the binary class case. In PMC-PBC-SVM algorithm, we divide a set of processors as a grid where each row is used to solve different binary case in parallel which means multi case is solved in parallel too, which means two level parallelisms.
The PMC-PBC-SVM algorithm combines Parallel Binary Class with parallel Multi Class Support Vector Machines for classification. It includes seven steps and works as follows:
- Reads a data set from input file
- Groups samples of the same class together
- Collects each two classes into one task
- Sort K(K-1)/2
-
Divides processes group as a grid where each row is used to solve different binary case in parallel at the same time, which means multi case is solved in parallel too
-
Builds SVM model after solving all binary tasks
-
Finally, write SVM model into an output model file which is used to predict the testing data set file.
Pmc-Pbc-Svm Results Analysis
The PMC-PBC-SVM algorithm is implemented using C++ programming language and its parallel version uses MPI2 communication routines 18. The experiments are carried out on an ALBACORE cluster19, which contains 12 compute nodes, each node has 2 chips, and each chip has 4 cores. Each core is an Intel(R) Xeon(R) CPU X5570 with 2.93 GHZ and 8192 KB Cache. Each node of node0 and node1 has 16 GB, and node2 to node11 has 12 GB. PMC-PBC-SVM has been tested using four different data sets with different sizes.
The first data set is the Earthworm data set20, which represents the small data set where its size has 248 of training samples, 3 classes, and 869 features. The second data set is the Protein data set21, which represents the medium data set where its size has 17766 of training samples, 3 classes, and 357 features. The third data set is the Mnist data set22, which represents large data set where its size has 60000 of training samples, 10 classes, and 780 features. The fourth data set is the Minst8m data set23, which represents the very large data set where its size has 8100000 of training samples, 10 classes, and 784 features.
The parallel performance is evaluated using the relative speedup (S), which is defined as the ratio of the time taken to solve a problem on a single processing element to the time required to
solve the same problem on a parallel computer with p identical processing elements, then L
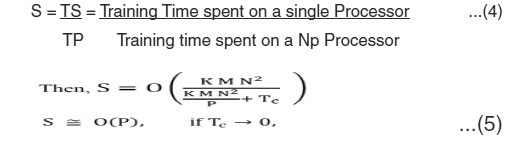
When transmitted data between processor in insignifian, In figure (2), the table shows the running time results of applying PMC-PBC-SVM implementation using different numbers of processors with different size data sets, Earthworm, Protein, Mnist, and Mnist8m, where real time multiply by 1000, 100, 10, and 1 respectively. The chart shows the relationship between run time and number of processors.
In figure (3), the table shows the speedup results of applying PMC-PBC-SVM implementation using different numbers of processors with different size data sets, Earthworm, Protein, Mnist, and Mnist8m. The chart shows the relationship between speedup and number of processors.
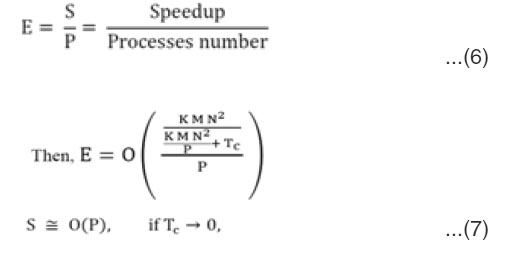
Efficiency (E) is another way to analyze the parallel implementation, which is defined as the ratio of speedup to the number of processing elements, then
When transmitted data between processor in insignfican, In figure (4), the table shows the efficiency results of applying PMC-PBC-SVM implementation using different numbers of processors with different size data sets, Earthworm, Protein, Mnist, and Mnist8m. The chart shows the relationship between efficiency and number of processors.
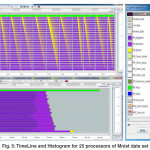
On the other hand, Jum pshot software 24, which is a visualization tool to study the performance
of parallel programs using log files that generated from the execution of the PMC-PBC-SVM implementation with different size data sets, Earthworm, Protein, Mnist, and Mnist8m. Figure (5) shows the outputs of Jumpshot which is the TimeLine and Histogram for 25 processors of Mnist data set.
From the results analysis, we can conclude that the PMC-PBC-SVM implementation is an efficient with small, medium and large data sets especially when data set has more than two classes. Regarding to very large data sets, the proposed algorithm works more efficiently. The results analysis shows that the PMC-PBC-SVM implementations can significantly improve the performance of data classification without sacrificing accuracy. The larger the data set is, the higher efficiency PMC-PBC-SVM is achieved.
Conclusion
In this paper, we set out to show that the parallel computing could be utilized in a way that would allow us to solve both binary and multi class classification problem of large data sets. In this research work, an efficient parallel algorithm, PMC-PBC-SVM, is developed. The proposed parallel algorithm combines Parallel Binary Class with Parallel Multi Class Support Vector Machines for classification. PMC-PBC-SVM algorithm was implemented using C++ and MPI. The parallel codes were executed on an ALBACORE Linux cluster. The parallel implementation was tested with four data sets with different sizes. Also, performance analysis was visualized using Jumpshot software. The results show that the PMC-PBC-SVM implementations can significantly improve the performance of data classification without sacrificing accuracy. The larger the data set is, the higher efficiency PMC-PBC-SVM is achieved.
In the future work, there will be a need for the training algorithms to generate the classification models in real time. Such a scenario needs the classification algorithm to efficiently use different parallel computing resources and distributed resources like Grid computing in an efficient manner. Since number of the class play an important role in deciding the effectiveness 8
of PMC-PBC-SVM. Therefore, there is a need for more efforts that can be developed to divide classification problem efficiently while maintaining accuracy, which is a good area for future work. The developed parallel algorithm, PMC-PBC-SVM, runs on both ALBACORE [24] Linux clusters and MCSR [25] supercomputers. The results are showed that the developed parallel algorithms work efficiently on Linux clusters more than supercomputers. Therefore, more future work can be doing in this area. On the other hand, a web based system can be build for the developed parallel algorithms as a future work.
Acknowledgement
The authors gratefully acknowledge the Northern Border University for their financially support and the University of Southern Mississippi for their scientifically support.
References
- Jiawei Han and Micheline Kamber. Data Mining: Concepts and Techniques. Book, Second Edition, 2006.
- C. Zhang, P. Li, A. Rajendran, and Y. Deng. Parallelization of multicategory support vector machines for classifying microarray data. BMC Bioinformatics, 2006.
- C. Cortes, and V. Vapnik. Support-vector networks. Machine Learning, 1995.
- Rajendran. Paralle Support Vector Machines for Mulicategory Classification of Large Scale Data. Dissertation, University of Southern Mississippi, 2007.
- Chih-Chung Change and Chih-Jen Len. LIBSVM: a library for support vector machines. Technical report, http://www.csie.ntu.edu.tw/_cjlin/libsvm/, 2005.
- J. Dong, A. Krzyzak, and C. Suen. Fast SVM training algorithm with decomposition on very large data sets. IEEE Transactions on Pattern Analysys and Machine Intelligence 27, pages 603-618, 2005.
CrossRef
- Thorsten Joachims. SVM Light, Support vector machine. Developed at the university of Dortmund , and it is available at http://svmlight.joachims.org/, 1994.
- G. Zanghirati, and L. Zanni. Parallel solver for large quadratic programs in training support vector machines. Parallel Computing, 29: 535-551, 2003.
CrossRef
- T. Serafini, G. Zanghirati, and L. Zanni. PGPDT: Parallel Gradient projection based on Decompostion Technique. Technical report, http://dm.unife.it/gpdt/,2007.
- Rabie Ahmed, Adel Ali, Chaoyang Zhang. SMC-PBC-SVM: A parallel implementation of Support Vector Machines for data classification. Conference on Parallel and Distributed Processing (PDPTA 2012).
- T.Eitrich, and B. Lang. Efficient Implementation of serial and parallel support vector machine training with a multi-parameter kernel for large-scale data mining. Proceeedings of World Academy of Science, Engineering, and Technology 11, 2006.
- S. Qiu, and T. Lane. Parallel computation of RBF kernels for support vector classifiers. Fifth SIAM International Conference on Data Mining, 2005.
CrossRef
- T. Serafini, L. Zanni. On the Working Set Selection in Gradient Projection-based Decomposition Techniques for Support Vector Machines. Optim. Meth. Soft. 20: 583-596, 2005.
CrossRef
- Rong-En Fan, Pai-Hsuen Chen, and Chih-Jen Lin. Working set selection using second order information for training support vector machines. Journal of Machine Learning, 8: Research, (2005).
- T.Eitrich, and B. Lange. On the optimal working set size in serial and parallel support vector machine learning with the decomposition algorithm. Proceedings of the fifth Australasian Conference on Data mining and analysis 61, pages 121-128(2006).
- J. Platt. Fasting training of support vector machines using Sequential Minimal Optimization. In Advances in Kernel Methods Support Vector Learning, MIT press, pages 185-208, 1999.
- J. Dong, A. Krzyzak, and C. Suen. A fast parallel optimization for training support vector machine. Proceedings of 3rd Int. Conf. Machine Learning and Data Mining, pages 996-105, Germany, 2003.
CrossRef
- Ewing Lusk, Steve Huss-Lederman. Message Passing Interface Forum. MPI: A Message-Passing Interface Standard (Version 2.2). September 4, 2009. URL http://www.mpi-forum.org/docs/mpi-2.2/mpi22-report.pdf.
- USM: Albacore Cluster, Chemistry lab at The University of Southern Miss. Technical report, http://albacore.st.usm.edu/cgi-bin/portal.cgi.
- Li Y, Wang N, Perkins EJ, Zhang C, Gong P. Identification and Optimization of Classifier Genes from Multi-Class Earthworm Microarray Data set. PLoS ONE 5(10): e13715. doi:10.1371/journal.pone.0013715, October 2010.
CrossRef
- Jung-Ying Wang. Application of support vector machines in bioinformatics. Master’s thesis, Department of Computer Science and Information Engineering, National Taiwan University, 2002.
- Yann LeCun, L. Bottou, Y. Bengio, and P. Haffner. Gradient-based learning applied to document recognition. Proceedings of the IEEE, 86(11):2278-2324, November 1998. MNIST database available at http://yann.lecun.com/exdb/mnist/.
CrossRef
- Gaëlle Loosli, Stéphane Canu, and Léon Bottou. Training invariant support vector machines using selective sampling. In Léon Bottou, Olivier Chapelle, Dennis DeCoste, and Jason Weston, editors, Large Scale Kernel Machines, pages 301-320. MIT Press, Cambridge, MA, 2007.
- Anthony Chan, David Ashton, Rusty Lusk, William Gropp. Jumpshot-4 Users Guide. Mathematics and Computer Science Division, Argonne National Laboratory. July 11, 2007.
- MCSR: Mississippi center for super computing research, Technical report, http://www.mcsr.olemiss.edu.

This work is licensed under a Creative Commons Attribution 4.0 International License.